Illing, Gerstner & Brea (2019) - Biologically Plausible Deep Learning - but how far can we go with shallow networks?
28 Jun 2021You can find the paper here.
Why am I reading this paper?
Stumbled upon this paper while researching how the delta rule is actually implemented in SNN: ex. which error signal do we consider? how to summarize the activity of the output layer over time into a single value to compute the loss?
Motivation
-
We (neuroscience/neuromorphic community 😀) care about the biological plausibility of the SNN learning rule mainly because of the current advances in neuromorphic hardware. People are looking for learning rules that can be implemented online, on chip…which leads us to local learning rules.
-
The question then becomes how far can we go with a simple model composed of one hidden layer in which only the readout layer is locally trained? I find it a particularly interesting question because the performance of such simple model can be used as benchmark against the more elaborate models out there. In a way, it can help draw a baseline on what SNN can do without any complex training procedures or architectures.
- So what is really out there for biologically-plausible training of SNN? i.e what are the alternatives to backprop for supervised training of multi-layer networks? Obviously there are two possibilities. For both cases, only the output (aka readout) layer is trained in a supervised fashion. Usually, this is a delta-rule. We can either:
- Fix the weights of the first layer to random values: ex. extreme learning machine
- Train the first layer(s) with unsupervised learning which is particularly appealing because can be implemented with local learning rules
- For completeness, more elaborate SNN models available in the literature include Conv layers with weight sharing, backprop approximations, multiple hidden layers, dendritic neurons, recurrence, conversion form rate to spikes.
Main Contribution
Interesting comparison study contrasting a simple yet promising model trained with biologically plausible local learning rule to deep learning models trained with backprop.
The main question the authors try to address is the following:
Given a single hidden layer network and a biologically-plausible, spike-based, local learning rule, how well can we perform on standard classification tasks compared to rate-based networks trained with backprop.
- Main outcome of this paper is that even a simple model trained with online stochastic GD (no minibatch) and a constant learning rate demonstrates comparable results to more “complicated” (aka elaborate) approaches.
Methods
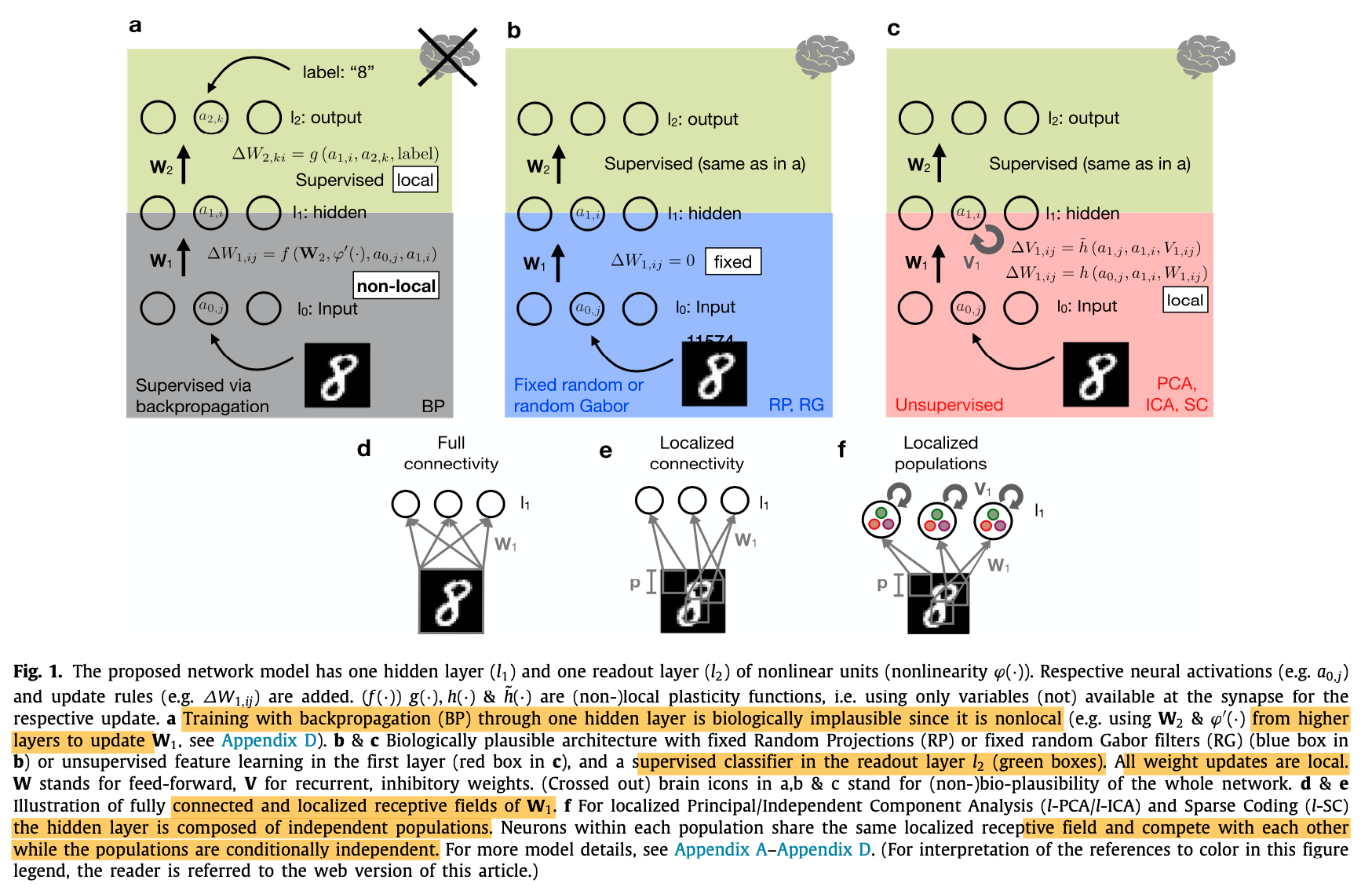
- Study different network topology and training procedure. All the studied networks consist of a single hidden layer:
- Network trained with backprop [rate-based]
- Network with fixed random projections (RP) or random Gabor filters (RG) [rate-based and spike-based]
- Network trained with unsupervised learning in the hidden layers (ex. PCA, ICA, Sparse Coding (SC)) [rate-based]
- Network with sparse connectivity between input and hidden:either localized connectivity where the input to hidden is not fully connected or localized populations in which case the hidden layer is composed of independent populations sharing the same receptive field but compete with each other. This is to force the hidden units to learn different features. [rate-based and spike-based]
- For the localized receptive field, this is a similar concept to CNN, a patch spanning p x p pixels of the input image is connected to particular neurons of the hidden layer.
- A simple perceptron (SP) without a hidden layer: i.e direct classification of the input, serving as a lower bound on performance [rate-based]
- Performance of these networks is compared on the MNIST task.
Local Supervised Learning Rule
The supervised learning rule used for SNN is a supervised delta-rule via STDP:
- Error signal computed as the difference between a post-synaptic spike-trace $tr_i(t)$ and a post-synaptic target trace $tgt_i(t)$.
- Readout weights $w_2$ are updated at every pre-synaptic spike time $t_j^f$.
- Target trace $tgt_i(t)$ is a constant and predefined.
-
Post-synaptic trace $tr_i(t)$ is updated at every post-synaptic spike time $t_i^f$.
$ \tau_{tr} \frac{dtr_i(t)}{dt} = -tr_i(t) + \sum_f \delta (t-t_i^f) $
$ \Delta w_{2,ij} = \alpha. (tgt_i^{post}(t) - tr_i^{post}(t)) \delta (t-t_j^f) $
Results
-
Unsupervised methods and random features projections perform better when paired with localized connectivity as compared to fully-connected topology.
-
Random fixed inout weights/ Gabor filters perform better than using unsupervised learning for the input layer. In other words, using unsupervised learning does not add performance advantage.
-
Using localized random projected and localized Gabor filters reach >98% accuracy on MNIST dataset which is comparable to other biologically plausible DL models
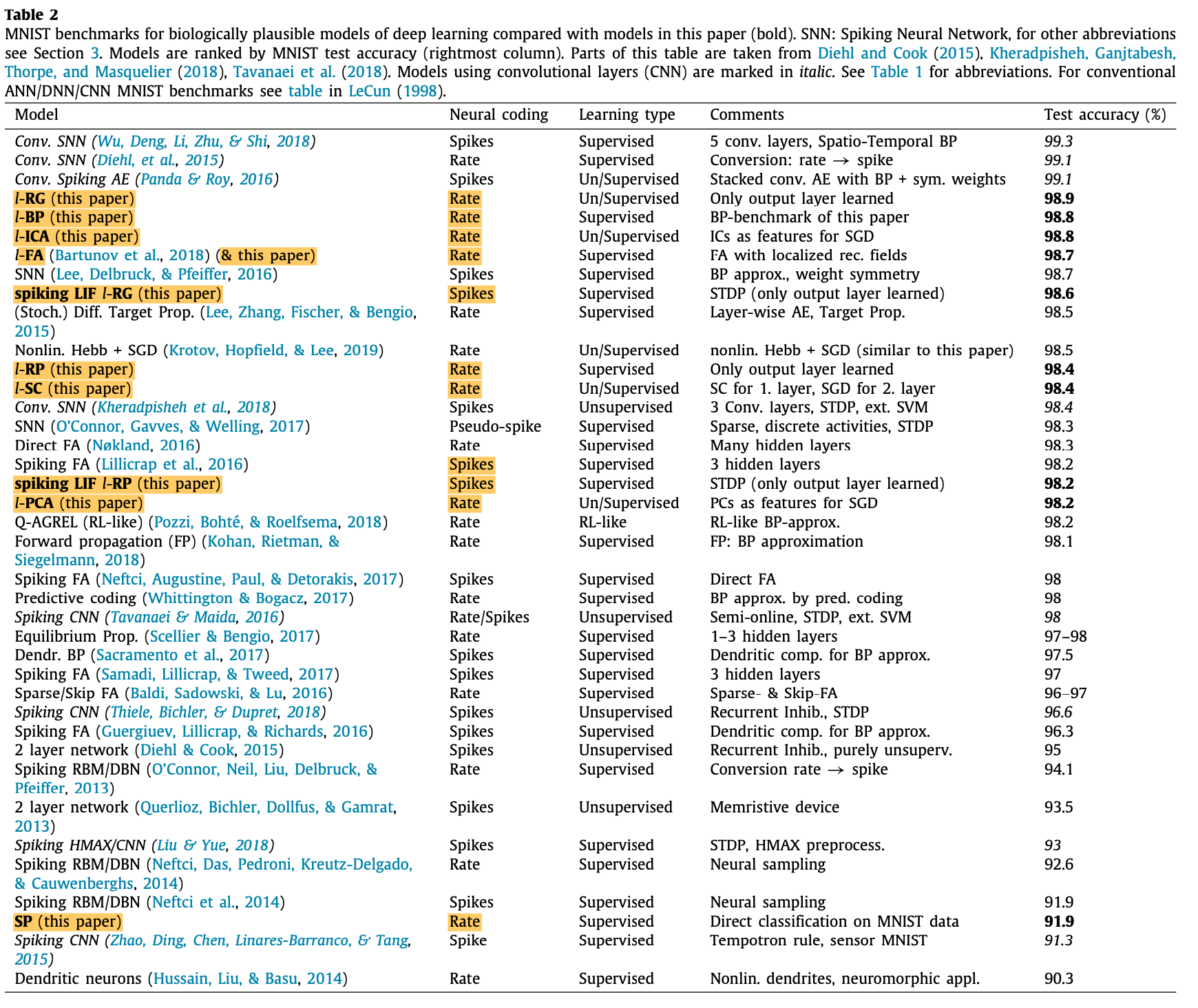
A summary of networks performance on MNIST for biologically plausible DL models can be found in the table below: